IOTA: Difference between revisions
(→Output) |
|||
| Line 211: | Line 211: | ||
= Output = | |||
Due to ''IOTA's'' flexibility, there are several types of output that co-exist simultaneously and can be somewhat disconnected from one another. It helps to think of them as three separate stages of the process: pre-processing, grid-search / integration, and post-processing / analysis. | Due to ''IOTA's'' flexibility, there are several types of output that co-exist simultaneously and can be somewhat disconnected from one another. It helps to think of them as three separate stages of the process: pre-processing, grid-search / integration, and post-processing / analysis. | ||
| Line 224: | Line 224: | ||
Finally, the "integration/###" folder itself contains text files with lists of images, e.g. input images, all integrated images, all images that failed integration, major clusters from the unit cell-clustering module, etc. The main logfile (iota.log) is also found here, as is the default input file for ''PRIME'' (prime.phil). | Finally, the "integration/###" folder itself contains text files with lists of images, e.g. input images, all integrated images, all images that failed integration, major clusters from the unit cell-clustering module, etc. The main logfile (iota.log) is also found here, as is the default input file for ''PRIME'' (prime.phil). | ||
== ''IOTA'' GUI Tutorial == | |||
=== Sample Dataset: Synaptic Fusion Complex === | |||
For the purposes of this tutorial we will use serial diffraction data collected from good ol' hen egg-white lysozyme (HEWL) crystals using synchrotron radiation. I know, I know, you're tired of lysozyme, but before we tackle the Grand Problems of Crystallography (tm), we'll use a good, reliable dataset to learn how to use this software. Onward! | |||
=== Step 1: Obtain the sample data === | |||
1. Make sure the ''cctbx.xfel'' is installed and available on your configuration. The software is available with the latest ''Phenix'' distribution. As ''cctbx.xfel'' is under active and vigorous development, make sure you install the latest ''Phenix'' nightly build to obtain the latest version of ''cctbx.xfel''. | |||
2. Create a new directory (e.g. 'iota_tutorial') in your user space, to ensure that you'll have read-write permissions. Go into that folder. From now on, all the files will be written there. | |||
3. Download the compressed tarball containing [http://smb.slac.stanford.edu/~templates/sample_serial_data/HEWL_synch_serial.tar.gz diffraction images]. | |||
4. Once the download is complete (it might take a while), create a subfolder in your iota_tutorial folder called images, move the tarball there and issue: | |||
gunzip HEWL_synch_serial.tar.gz | |||
and then | |||
tar -xvf HEWL_synch_serial.tar | |||
NOTE: If you explore the folder, you will notice that the images are located in a somewhat complex tree of subfolders; the subfolders ./365/106 and ./365/406 refer to the two cryo-cassettes which contained the frozen crystals, while the subfolders at the next level represent the cassette positions for each crystal. Anywhere between 2 and 20 images were collected from each crystal, so the number of images per subfolder varies significantly. In general, it's best to run IOTA "alongside" the data folder, and to not point IOTA to a folder that can contain both raw and processed images, or images that you do not wish to process, as IOTA will find and attempt to process them all, causing problems. | |||
'''[UNDER CONSTRUCTION!]''' | |||
Revision as of 22:14, 16 May 2018
IOTA: integration optimization, triage and analysis
IOTA is a user-friendly front end for the cctbx.xfeland DIALS suites of serial diffraction data processing programs. It is comprised of three main modules:
- Raw image import, conversion, pre-processing and triage
- Image indexing and integration using cctbx.xfel (with optimization of spot-finding parameters) or DIALS (this is currently in the process of being adapted for diffraction stills)
- Analysis of the integrated dataset
IOTA can be run as a GUI or from the command-line; scripts can be used for both, interchangeably. The GUI has the advantage of displaying useful statistics; it can also be run in "monitor mode" during live data collection, during which the program will wait for new images to be written into the specified input folder. The command-line mode is useful if the program is run remotely on servers that do not, for some reason, support graphics.
Please note that IOTA is a front-end for (currently) two pieces of data processing software: cctbx.xfel and DIALS. Therefore, the preferred construction for citation should be something like "diffraction data were processed with IOTA [1] using data reduction algorithms implemented in cctbx.xfel [2] (or DIALS [3])".
[1] IOTA: integration optimization, triage and analysis tool for the processing of XFEL diffraction images. Lyubimov AY, Uervirojnangkoorn M, Zeldin OB, Brewster AS, Murray TD, Sauter NK, Berger JM, Weis WI, Brunger AT. J Appl Crystallogr. 2016 May 11;49(Pt 3):1057-1064
[2] Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers. Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers. Hattne J, Echols N, Tran R, Kern J, Gildea RJ, Brewster AS, Alonso-Mori R, Glöckner C, Hellmich J, Laksmono H, Sierra RG, Lassalle-Kaiser B, Lampe A, Han G, Gul S, DiFiore D, Milathianaki D, Fry AR, Miahnahri A, White WE, Schafer DW, Seibert MM, Koglin JE, Sokaras D, Weng TC, Sellberg J, Latimer MJ, Glatzel P, Zwart PH, Grosse-Kunstleve RW, Bogan MJ, Messerschmidt M, Williams GJ, Boutet S, Messinger J, Zouni A, Yano J, Bergmann U, Yachandra VK, Adams PD, Sauter NK. Nat Methods. 2014 May;11(5):545-8.
[3] Diffraction-geometry refinement in the DIALS framework. Waterman DG, Winter G, Gildea RJ, Parkhurst JM, Brewster AS, Sauter NK, Evans G. Acta Crystallogr D Struct Biol. 2016 Apr;72(Pt 4):558-75.
IOTA GUI
The most user-friendly way to run IOTA is in GUI mode. This starts up simply by issuing
iota
As a shortcut, IOTA GUI can be launched with an existing script supplied as a command-line argument, like this
iota iota.param
If that is done, the elements of IOTA GUI will be populated with the parameters specified in the script. Also available as command-line arguments: the path to the data folder / file, turn on monitor mode, supply the number of processors for the multiprocessing run. New options are added all the time; check which options may be available by issuing
iota -h
Main Window
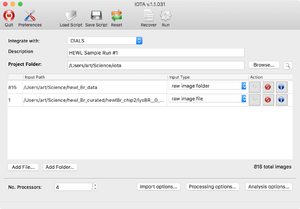
First you will see the main input screen, which will allow you to enter basic information, such as the output folder and the project description. The input file list has "Add Folder" and "Add File" buttons, which allow you to input multiple sources of data: individual diffraction images, folders with diffraction images (or subfolders, etc.), and text files containing lists of paths to diffraction images (absolute paths work best here). As each item is added, a line is generated showing the number of images therein, as well as "actions" that can be taken. The entry can be deleted from the list (middle button), or the images can be viewed using an image viewer (left button, with a diffraction icon). "IOTA" would launch the image viewer appropriate to the backend selected from the "Integrate with" dropdown menu; thus either "DIALS" or "cctbx" image viewer would open.
As entries are added, a total number of read-in images is reported in the lower right corner. Once all the inputs are read in, the user can customize their IOTA run by changing the various preferences and options.
Settings
GUI Preferences
The Preferences toolbar button opens a dialog which allows the user to set some settings for the IOTA GUI, among them the choice of the multiprocessing method, two ways to select a subset of images (a set of image ranges and/or a random selection, these are compatible), monitor mode options, etc.
Currently, three multiprocessing modes are available (by clicking on the "Preferences" toolbar button): 'multiprocessing' refers to merely using multiple cores on your local machine, 'lsf' will allow you to submit jobs to an LSF queue, while 'torq' refers to the queue set up at SSRL's processing servers (this one is under construction). The queues can be selected from a drop-down list or, if not found on the list, a queue name can be supplied by user.
Processing Options
The main screen contains three buttons that open dialogs for image import options, processing options (this varies depending on backend choice) and analysis options. The image import options dialog allows the user to turn on/off image triage (i.e. image rejection based on whether sufficient Bragg spots are found), override beam XY and detector Z coordinates, threshold out the beamstop shadow, specify a mask, etc. NOTE: as the serial data processing algorithms become more sophisticated, and the equipment at synchrotron and XFEL facilities is optimized for serial data collection, most of these overrides are becoming unnecessary. However, supplying a good beamstop shadow mask is still a very useful step. Users can do this in the DIALS image viewer, which can be launched either independently, or by clicking on the "view image" button associated with an entry in the Input Window.
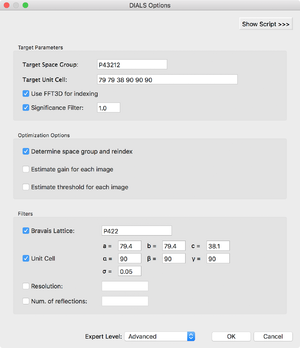
The processing options dialog allows the user to generate a default target (PHIL) file for cctbx.xfel / DIALS or read in an existing one, and modify the settings manually in a text window (revealed by clicking the "Show Script" button). Furthermore, you can modify spot-finding grid search options and integration result filter options. We have tried to supply reasonable default presets for both backends; however, optimization of data processing algorithms is still a work in progress and some users may find it necessary to play with some of the more obscure parameters. At the moment, we encourage users to contact IOTA developers with any questions.
Analysis Options
The analysis dialog allows you to output various charts summarizing IOTA output as well as individual image integration results. Most of the charts are a remnant of the older, command-line version of IOTA; they have been superseded by the charts shown in the run-time GUI. However, users who desire an in-depth, image-by-image look into their data processing, can turn on these features. They include: 1. Integration predictions overlayed on diffraction images; 2. Plots of lattice model shifts during refinement; 3. Mosaicity "trumpet" plots. Most of these are generated by the cctbx.xfel and DIALS backends. WARNING: The generation of these plots may slow down your IOTA run!
Also note: the unit cell clustering option is off by default, as the module seems to conflict with some installations of the cctbx suite of software. If the user doesn't turn the clustering option on, it can be initiated after the processing run is concluded (see below).
Run Statistics and Analysis
Once IOTA is running, a run-time processing window will appear with two tabs: a Log tab that will display iota.log as it is updated in real time, and the Charts tab, which will display several useful graphs: of resolution vs. frame, number of strong (I / sigI > threshold) spots per frame, a histogram of unit cell parameters, a plot of indices with measurements, and a bar chart breaking down indexing / integration success for the full dataset. The processing window will also allow the user to turn on the "Monitor Mode", in which IOTA will continuously check if any new diffraction images have been added to the input folder (or subfolders therein); this is a useful mode to use when running IOTA concurrently with data collection.
The log text is searchable, allowing the user to see the log entry for any specific image. Several of the charts are clickable: the resolution / number of spots charts allow the user to click on any individual point on the scatter plot, learn the associated filename, and launch DIALS image viewer to view the image; the plot of indices can be clicked to view a h=0, k=0, or l=0 slice; a double-click on any segment of the run summary plot will allow the user to view all or some of the images associated with that particular group (e.g. if the user double-clicks on the 'Failed Indexing' fraction, they can then view all or a portion of the images that could not be indexed).
When the run finishes, a new Analysis tab will appear in the processing window. There, the pertinent summary of the run would be displayed, along with buttons that will display several useful charts: a heatmap of the spot-finding results (if the cctbx.xfel backend was used), resolution histograms and beam XYZ charts. The user can run unit cell clustering from this window (results will be displayed in the table) with different options, if desired. The user can also choose to run PRIME from this window, in which case the PRIME GUI will launch with the parameters pertinent to this run filled in (e.g. input / output folders, resolution limits, pixel size, unit cell, etc.)
IOTA in Command Line
Auto Mode
The simplest way to run IOTA is in Auto Mode. To do so, simply issue:
iota.run /path/to/image/files/
Alternatively, if a text file with a list of images exists, IOTA can accept that file as input:
iota.run input_images.lst
Once running, IOTA will display a program logo, some information about the configuration of the run and a progress bar for each major step, e.g.:
-bash-4.1$ iota.run ../hewl_Br_data
IIIIII OOOOOOO TTTTTTTTTT A
II O O TT A A
II O O TT A A
>------INTEGRATION----OPTIMIZATION--------TRIAGE-------ANALYSIS------------>
II O O TT A A
II O O TT A A
IIIIII OOOOOOO TT A A v1.1.031
with DIALS
Monday, May 14, 2018. 09:51 AM
IOTA will run in AUTO mode using /Users/art/Science/hewl_Br_data:
Reading input files -- DONE................................................0.65s
IMPORTING IMAGES: 5% [ --- ] [ ==> ]
IOTA will automatically create two script files: iota.param (which contains settings for running IOTA) and cctbx.phil or dials.phil (a cctbx.xfel or DIALS target file), which can be modified by a user to fine-tune various settings, or read into IOTA GUI if desired. The output will be collected in the folder named "integration", which will contain subfolders for each integration run, titled "001", "002", "003", etc. Each run generates a folder named "final" with the final integrated pickles as well as individual cctbx.xfel logs for each image. Furthermore, lists of files that have been successfully integrated (integrated.lst), failed integration (not_integrated.lst), etc. can be found there. Finally, a pre-populated script for PRIME (prime.phil) can be found there as well. (Currently, the user must manually edit prime.phil to specify the number of residues - "n_residues" - in order to run PRIME successfully.)
Target Files
IOTA itself is a front-end to the data processing programs cctbx.xfel and DIALS. These programs require their own set of parameters, distinct from IOTA parameters, which are located in so-called "target" files: text files containing parameters encoded in Python-based hierarchical interchange language or PHIL. When run in AUTO mode, IOTA generates an appropriate target file for cctbx.xfel or DIALS using defaults deemed reasonable for most serial crystallography projects. These default target files can also serve as a starting point for the user to modify those settings as they see fit. The user has the option to provide their own target file (perhaps generated during a previous data processing attempt). The user can edit the IOTA settings to specify the target file
cctbx {
target = "dials.phil"
}
or use a command-line argument
iota.run /path/to/image/files/ dials.target=user_params.phil
Script Mode
IOTA can be run using a script file, e.g.:
iota.run script.param
The script contains settings in PHIL format, e.g.:
description = "IOTA run #5, with some modified settings"
input = "/path/to/raw/images/"
output = "/path/to/iota/output/"
image_conversion {
rename_pickle = None keep_file_structure *auto_filename custom_filename
rename_pickle_prefix = None
convert_only = False
square_mode = None no_modification *pad crop
mask = None
invert_boolean_mask = False
beamstop = 0
distance = 0
beam_center {
x = 0
y = 0
}
}
image_triage {
type = None no_triage *simple grid_search
min_Bragg_peaks = 10
grid_search {
area_min = 6
area_max = 24
height_min = 2
height_max = 20
step_size = 4
}
}
.
.
.
The script can be auto-generated (with an accompanying target.phil file with some default cctbx.xfel settings) via a "dry run" by issuing
iota.run -d
The same "-d" command-line option will print to terminal the full IOTA script file with help statements.
Additionally, IOTA settings can be modified by command-line statements, e.g.:
iota.run script.param cctbx.grid_search.type=smart cctbx.grid_search.area_median=7
Single-Image Mode
IOTA can accept a single image as input:
iota.run images/img_00001.pickle
Alternatively, IOTA can be run in bare-bones "single-image mode"
iota.single_image images/img_00001.pickle
These options are best for testing purposes.
Command-line Options
In addition to a command script, IOTA runs can be modified by command-line options:
-h, --help show help message and exit --version Prints version info -l, --list Output a file (input_images.lst) with input image paths and exit -c, --convert Convert raw images to pickles and exit -d, --default Generate default iota.param and target.phil files and exit -p PREFIX, --prefix PREFIX Specify custom prefix for converted pickles (e.g. -p user) -s, --select Selection only, no grid search -r RANDOM Run IOTA with a random subset of images, e.g. "-r 5" -n NPROC Specify a number of cores for a multiprocessor run" --mpi [MPI] Specify stage of process - for MPI only --analyze [ANALYZE] Use for analysis only; specify run number or folder with results
These options can be shown by issuing:
iota.run -h
Perhaps the most useful of these are -r and -n options, as they allow the user to adjust an IOTA run in Auto-mode on the fly. Alternatively, both of these settings can be changed within the script file.
All of the options in the script can be introduced as command-line statements by using a "compressed" PHIL format. Thus:
cctbx {
grid_search {
type = None *brute_force smart
}
}
translates into
iota.run script.param cctbx.grid_search.type=brute_force
Output
Due to IOTA's flexibility, there are several types of output that co-exist simultaneously and can be somewhat disconnected from one another. It helps to think of them as three separate stages of the process: pre-processing, grid-search / integration, and post-processing / analysis.
In pre-processing, raw images are read in and converted to Python pickles. These are saved under the "converted_pickles" folder in the format <prefix>_<run_no>_<#####>.pickle; each cycle of pre-processing is assigned a run number (e.g. "001", "002", "003", etc.). Pre-processing is only triggered if a) the read-in image is not already pickled or b) the image has to be modified in some way (e.g. override beamXY coordinates, change detector distance, etc.). Thus, if converted and modified pickles are submitted to IOTA, the "converted_pickles" folder will not be created. The purpose of this is to allow the user to experiment with image modification, then subsequently select the converted pickles that best fit the user's needs.
The output of the other two steps (grid-search / integration and post-processing / analysis) can be found under the "integration" folder. The grid-search results are saved to the "integration/###/image_objects" folder in the format <filename>.int. These are pickled dictionaries which contain all the information about the individual images (without the pixel values or integrated intensities), such as raw image filename, converted pickle filename, the details of the grid search, etc. These can be used for some of the advanced operations, such as experimentation with the selection process without repeating the grid search.
The integrated pickles are collected under "integration/###/final" folder, in the format int_<filename>.pickle. Only successfully integrated images are saved this way. For each of the input images, however, a log of cctbx.xfel or DIALS output is saved in the same folder, in the format <filename>.log. This log documents each integration attempt from the grid-search with the final integration attempt at the end (for cctbx.xfel) or the linear indexing/integration output (for DIALS) and can be used for troubleshooting.
If the user chooses to output any charts (e.g. grid-search heatmap, beam center plot, image visualization, etc.), these will be found under "integration/###/visualization" folder.
Finally, the "integration/###" folder itself contains text files with lists of images, e.g. input images, all integrated images, all images that failed integration, major clusters from the unit cell-clustering module, etc. The main logfile (iota.log) is also found here, as is the default input file for PRIME (prime.phil).
IOTA GUI Tutorial
Sample Dataset: Synaptic Fusion Complex
For the purposes of this tutorial we will use serial diffraction data collected from good ol' hen egg-white lysozyme (HEWL) crystals using synchrotron radiation. I know, I know, you're tired of lysozyme, but before we tackle the Grand Problems of Crystallography (tm), we'll use a good, reliable dataset to learn how to use this software. Onward!
Step 1: Obtain the sample data
1. Make sure the cctbx.xfel is installed and available on your configuration. The software is available with the latest Phenix distribution. As cctbx.xfel is under active and vigorous development, make sure you install the latest Phenix nightly build to obtain the latest version of cctbx.xfel.
2. Create a new directory (e.g. 'iota_tutorial') in your user space, to ensure that you'll have read-write permissions. Go into that folder. From now on, all the files will be written there.
3. Download the compressed tarball containing diffraction images.
4. Once the download is complete (it might take a while), create a subfolder in your iota_tutorial folder called images, move the tarball there and issue:
gunzip HEWL_synch_serial.tar.gz
and then
tar -xvf HEWL_synch_serial.tar
NOTE: If you explore the folder, you will notice that the images are located in a somewhat complex tree of subfolders; the subfolders ./365/106 and ./365/406 refer to the two cryo-cassettes which contained the frozen crystals, while the subfolders at the next level represent the cassette positions for each crystal. Anywhere between 2 and 20 images were collected from each crystal, so the number of images per subfolder varies significantly. In general, it's best to run IOTA "alongside" the data folder, and to not point IOTA to a folder that can contain both raw and processed images, or images that you do not wish to process, as IOTA will find and attempt to process them all, causing problems.
[UNDER CONSTRUCTION!]